Here is the link ⬇⬇⬇
click here

More info below ⬇️
click here



Find it here
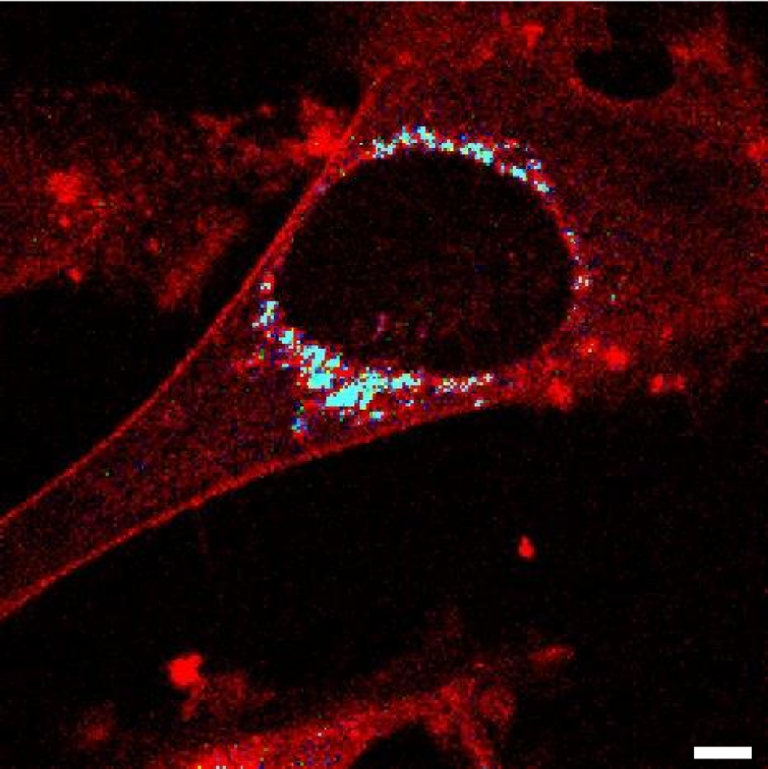
Researchers from our group (A. Karpińska, K. Kwapiszewska, J. Michalski, and R. Hołyst) performed single-molecule measurements proving successful intracellular delivery of the conjugates. We are happy that our original method for quantitative biology found its application in such an ambitious project.
The FCS-based method was developed within the LIDER/10/0033/L-9/17/NCBR/2018 project financed by The National Centre for Research and Development.
The article is here

The Polish Agency for Enterprise Development (PARP) organizes this prestigious competition under the auspices of the two ministries: the Ministry of Economic Development, Labour and Technology, and the Ministry of Education and Science.
SERSitive offers substrates for surface-enhanced Raman spectroscopy (SERS) that allow for fast and accurate analysis that found its way in various analytical sciences such as pharmacy, forensic laboratories, border guard laboratories, medicine, and many others.
You can read more here


The headquarter of a eukaryotic cell is the nucleus, and most of the cell’s information and instructions are stored there in the form of DNA (Deoxyribonucleic acid). The DNA, which is twisted, rolled and bundled two-meter-long chain, together with protein molecules, makes up the chromatin fiber that lays inside the nucleus. For years, scientists were curious how these components are organized. How is it possible that proteins necessary in biochemical reactions move efficiently within the nucleus full of DNA? Recent studies have finally solved the mystery. Findings describing it in detail were published in the Journal of Physical Chemistry Letters on December 21st, 2020.
Molecules in a crowded nucleus
The nucleus of each cell hides a two-meter-long chain of a most amazing and unique molecule: DNA. Along with histones and various related proteins, DNA builds up a chromatin framework filled with a viscous fluid that exhibits excellent molecular composition diversity. For decades, the mobility of molecules in the nucleus was not sufficiently explored, but recent developments have altered this status quo. Thanks to in-depth research by a group of researchers from the Institute of Physical Chemistry of the Polish Academy of Sciences (IPC PAS) led by professor Robert Hołyst, the mobility of molecules at length-scales from single to tens of nanometers in the nucleus is presented in detail.
A Molecular superstore
Due to its small size, one might assume that the nucleus has a simple structure and a random molecule distribution. That is by no means the case. The nucleus has an unbelievably complex and fine-tuned layout. The DNA does not resemble a messy tangle of spaghetti; it is efficiently packed into compact structures. Even the nanoscale viscosity of the nucleus determines the mobility of the individual objects inside. To better visualize how well-organized this all is, the nucleus can be described as a superstore. The chromatin fibers work like shelves, holding an assortment of necessary genetic information (i.e., DNA) just like the shop shelves are filled with products. These shelves don’t take up the entire space, but rather they are separated within an aisle-like distance that works as a channel. The people crossing the aisles in specific patterns as they shop could be compared to the protein molecules that move somewhat randomly within the nucleus’ channels according to the rules of Brownian motion. No matter how crowded the aisle gets, people always find a way to pass by each other, maintaining some distance as they go. The molecules crossing molecular channels do the same without any traffic problems on their way. This allows each molecule to travel efficiently, maintaining the orderliness of a superstore.
Viscosity impact
The molecules that are present in the eukaryotic cells have different sizes. For example, ions are subnanometer in size, protein radii are typically few nanometers; a nucleosome’s radius is about 5.5 nm, while folded up chromatin fibers have a radius of about 15 nm. Furthermore, condensed loops of chromatin form higher-level compact structures are boasting a radius of about 150 nm. To understand their mobility within the nucleus, professor Hołyst’s team proposed to place nanometer-sized objects covering the whole spectrum of natural components length-scales found in the nucleus. Polymers, proteins, and nanoparticles having radius from 1.3 to 86 nm in were considered.
To see this intriguing organization at the nanoscale level, specific molecules’ mobility was studied using non-invasive techniques such as fluorescence correlation spectroscopy (FCS) and raster image correlation spectroscopy (RICS). Thanks to substances like the GFP (green fluorescent protein) or the rhodamine-based nanoparticles in nanomolar concentration, it was possible to observe the mobility of particular molecules and determine the nucleoplasm viscosity without causing any disruption to cellular activity. These techniques allow scientists to investigate even the most minor changes at the molecular level. The mobility of large nanoparticles was reduced by as much as 6 times compared to the diffusion in an aqueous medium. However, the typical protein-sized molecules diffusion was reduced only 2-3 times. The mobility drastically decreases when the radius of injected objects is larger than 20, more significance on estimations of the diffusion coefficient, it is possible to look closer at the movement and interaction of molecules that happen between particular objects in the nucleus’ channels and within the packed structure inside the nucleus. These measurements expand our current understanding of the structure of the nucleus. Having a good understanding of the complexity of the channels within nuclei is crucial as it directly contributes to our knowledge of how large biostructures, perhaps including the medicine of the near future, are transported within the cell.
The first author, dr. Grzegorz Bubak remarks, “Our experiments revealed that eukaryotic cell nucleus is percolated by ∼150 nm-wide interchromosomal channels filled with the aqueous diluted protein solution of low viscosity.”
The studies quantifying the crowding within cells’ nuclei reveal that most molecules can freely pass through this complex structure. Based on experiments supported by theoretical models, it was possible to estimate channels’ width (~150 nm) between the chromatin structure. The nuclei channels can constitute as much as 34% of the nuclei’s volume which is around 240 fL. If they were narrower, the chromatin fibers would be more dispersed, making the molecules’ efficient movement inside impossible. It is fascinating that the nucleus can contain such large amounts of DNA and other chemical elements without disturbing the molecules’ migration. This is all thanks to the well-arranged chromatin fibers made by DNA with structural proteins that give the double helix its shape. The mobility of particular chemical elements through the biological fluid in molecular channels is essential in many processes, such as creating specific molecules and forming new protein complex structures.
“These results can be of great importance when designing biological drugs such as therapeutic proteins, enzymes, and monoclonal antibodies, which can have the hydrodynamic radii larger than conventional chemical drugs based on synthetic compounds.” – concludes dr. Bubak
As a result of these studies, the mobility of the molecules in the nuclear channels is now described in detail and well-understood for the first time. Thanks to the research presented in this work, we now know how the chromatin fibers govern molecule organization, revealing the intriguing molecular machinery hidden deep inside the nucleus. We are now one step closer to developing therapeutic agents that can be effectively transported into the nucleus.
This work was supported by the National Science Centre, Poland, within the grant Maestro UMO-2016/22/A/ST4/00017.
SCIENTIFIC PAPERS:
“Quantifying Nanoscale Viscosity and Structures of Living Cells Nucleus from Mobility Measurements”
Grzegorz Bubak, Karina Kwapiszewska, Tomasz Kalwarczyk, Krzysztof Bielec, Tomasz Andryszewski, Michalina Iwan, Szymon Bubak, Robert Hołyst
The Journal of Physical Chemistry Letters 12, 294-301 (2021)
DOI: 10.1021/acs.jpclett.0c03052
Author: Dr Magdalena Osial
Contact: mosial@chem.uw.edu.pl
